Projects
|
Projects |
|
|

Langmuir Front Cover Goswami V, Tomar VR, Yashika, Deep S (2024) "Nanocarriers for the Delivery of Quercetin to Inhibit the UV-Induced Aggregation of γD-Crystallin" Langmuir, 40, 5617-5631 |
Understanding the mechanism of protein aggregation.
Our laboratory has extensively studied aggregation of model proteins as well as neurodegenerative disease linked proteins using various biophysical techniques and MD simulations. These studies have contributed significantly to molecular level understanding of protein aggregation/ fibrillation. The group has delineated factors which can affect (a) a protein aggregating via nucleated versus downhill polymerization (b) a protein undergoing aggregation via fragmentation versus heterogeneous nucleation. Our group has identified polyphenols which can inhibit in vitro fibrillation of superoxide dismutase linked to ALS and break the fibrils. The group has also looked in detail the potential of polyphenols as inhibitor for formation of Parkinson linked alpha-synuclein fibrils formation, Alzheimer related Abeta fibril and Cataract related crystallin. Our group has diligently endeavored to unravel the nuanced mechanisms underlying the action of various polyphenols on fibril of a given protein. These compounds exhibit a spectrum of behaviors across different stages of the aggregation pathway. While certain polyphenols solely inhibit aggregation, others display the remarkable ability to dismantle preformed fibrils. Moreover, their effects vary across distinct protein targets. Notably, a specific polyphenol can fragment fibrils of one protein into smaller species, while inducing an amorphous morphology during aggregation of another protein.

(Human Seru Albumin) |
(Super Oxide Dismutase) |
(Bovine Carbonic Anhydrase) |
(Lysozyme) |
Sharma S., Deep S. (2024), "Inhibition of Fibril Formation by Polyphenols: Molecular Mechanisms, Challenges, and Prospective Solutions", Chem Comm, (Accepted)
Goswami V, Tomar VR, Yashika, Deep S (2024), "Nanocarriers for the Delivery of Quercetin to Inhibit the UV-Induced Aggregation of γD-Crystallin", Langmuir, 40, 5617-5631
Sharma S, Tomar VR, Deep S (2023), "Myricetin: A Potent Anti-Amyloidogenic Polyphenol against Superoxide Dismutase 1 Aggregation", ACS Chem Neurosci, 14, 2461–2475
Sharma S, Tomar VR, Jayaraj A, Deep S (2023), "A Computational Strategy for the Therapeutic Development Against Superoxide Dismutase (SOD1) Amyloid Formation: Effect of Polyphenols on the various events in the aggregation pathway", Phys Chem Chem Phys , 25, 6232-6246
Bhatia N.K., Modi P., Sharma S., Deep S. (2020), "Quercetin and baicalein act as a potent anti-amyloidogenic and fibril destabilizing agents for SOD1 fibrils", ACS Chem Neurosci, 11, 1129-1138
Zaidi F.,Deep S. (2020), "Scutellarin Inhibits the Uninduced and Metal-Induced Aggregation of ɑ-Synuclein and Disaggregates Preformed Fibrils: Implications for Parkinson's disease", Biochem J, 477, 645-670
Katyal N., Agarwal M., Sen R., Kumar V.,Deep S.(2018), " Paradoxical Effect of Trehalose on the Aggregation of α-Synuclein:Expedites Onset of Aggregation yet Reduces Fibril Load ", ACS Chem NeuroSci, 9, 1477–1491
Saha S., Deep S.(2016), " Glycerol Inhibits the Primary Pathways and Transforms the Secondary Pathway of Insulin Aggregation ", Phys Chem Chem Phys, 18, 18934-18948
Bhatia N.K., Srivastava A, Katyal N., Jain N.,Khan M.A.I.,Kundu B. Deep S.(2015), " Curcumin binds to the pre-fibrillar aggregates of Cu/Zn superoxide dismutase (SOD1) and alters amyloidogenic pathway resulting in reduced cytotoxicity", Biochim. Biophys. Acta, 1854, 426-436
Saha S., Deep S. (2014), " Switch in the Aggregation Pathway of Bovine Serum Albumin Mediated by Electrostatic Interactions" , J Phys Chem B, 118,9155–9166
Investigations of the interaction of transforming growth factor protein with receptor II and its mutants.
Transforming growth factors (TGF-b) (referred as ligands) are proteins that act as signaling molecules between cells. They regulate the differentiation and maturation of the target cells by binding to specific receptors, first to receptor II (TbRII) and then to receptor I (TbRI), on the surface of these cells. Mutations in these ligands/receptors have been implicated in a variety of human diseases including cancer. The structural data indicate that receptor signaling complexes with different isoforms of transforming growth factor are not significantly different, suggesting that they are functionally redundant. However, functional differences have been observed in the context of cutaneous wound healing, where the application of TGF-b3 prevents scarring, while addition of TGF-b1 promotes scarring. Also, the fact that the differences between TGF-? isoforms persist across the time span of vertebrate evolution argues against functional redundancy of these isoforms. Answer probably lies in the detail of the interactions. Two ligands may recognize the overlapping sites on the same receptor, but may be dependent on distinct residues for high affinity binding. Thus, it is important to identify the critical and distinct residues at the ligand-receptor interface. For this, we are investigating differential thermodynamic profiles of TbRII interaction to wild and interfacial residues variants of TGF-b3.
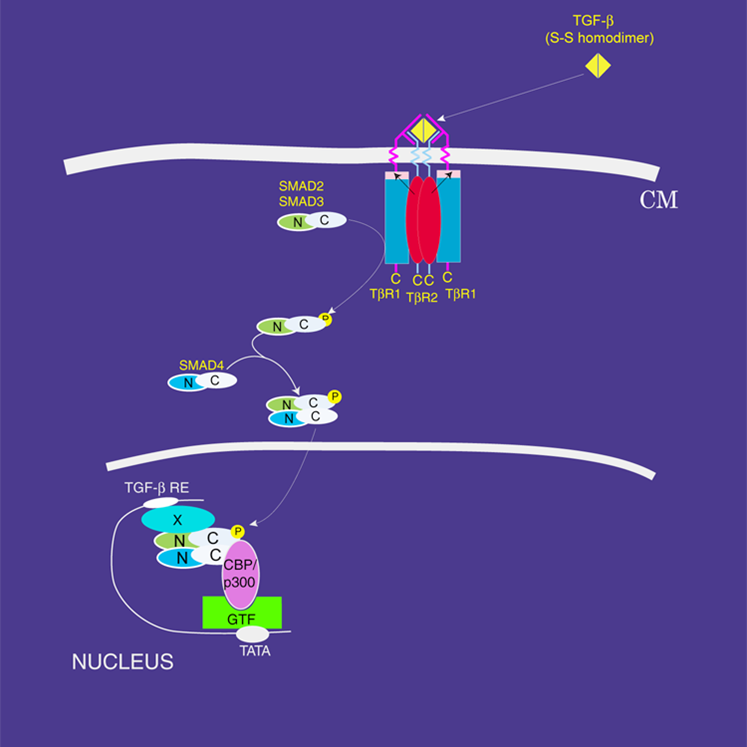
(Signal Transduction pathway of TGF-b3) |
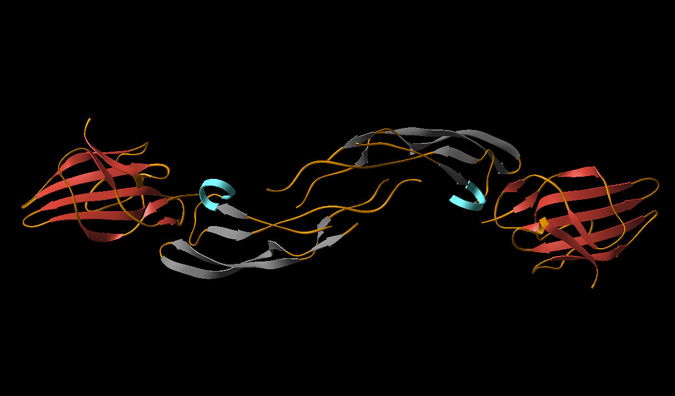
(TGF-b3 TbR-II complex) |
Dawn A,Deep S. (2021), "An Improved strategy of TGFβ3 Expression in Escherichia coli: exploiting the folding modulators for a switch from misfolded to folded form", Int J Biol Macromol, 167, 787-795
Dawn A, Khatri K.S., Karmakar S.Deep S. (2020), "Interaction of TGFβ3 ligand with its receptors type II (TβRII) and type I (TβRI): A unique mechanism of protein-protein association", Biochim Biophys Acta 1868, 140485
Dawn A.,Deep S. (2020), "Thinking beyond tradition: Polyphenols as effective refolding modulators", International Journal of Biological Macromolecules, 148, 969-978
Nayeem S.M., Oteri F., Baaden M.,Deep S.(2017), " Residues of Alpha Helix H3 Determine Distinctive Features of Transforming Growth Factor β3 ", J Phys Chem, 121, 5483-5498
Nayeem S.M.,Deep S.(2014), " pH modulates the TGF-ß ligands binding to the receptors: A computational analysis", Journal of Molecular Recognition, 27, 471-481
Nayeem S. M., Deep S. (2010), “Rationalization of poor solubility of TGF-b3 using MD simulation”. Biochemical Biophysical Research communication, 401, 544-547.
WhiB proteins as new drug target for Tuberculosis
Recently, Our group has ventured to look at new targets for Tuberculosis. Tuberculosis is the major cause of mortality in all over the world. Many fundamental questions about the mechanism used by M. tuberculosis for survival during early infection and persistence in the host cell is still unexplored. The modulation of its environment and the ability to enter a dormant phase are the hallmarks of M. tuberculosis. Since WhiB and Cas proteins are involved in many crucial functions of M. tuberculosis, so, their biochemical and biophysical properties needs to be explored further. It will help in identifying novel drug targets for Mycobacterium tuberculosis. Our lab has made effort in understanding the role of these proteins and using them as potential drug target.