
|
| |
  |
|
| About ZiF-Predict |
Background
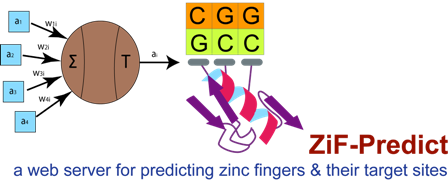
Zinc fingers are proteins containing a finger-shaped fold that coordinates zinc ions with a combination of cysteine and histidine residues for their fold stabilization. Their structure comprises of two anti-parallel β-sheets and an α-helix. Each finger binds to a DNA template in the major groove. Changing the amino acids at certain key positions generates zinc finger motifs with different triplet sequences and varying binding specificities. Our algorithm considers the positions -1, +3 and +6 relative to the start of the zinc finger protein (ZFP) α-helix (key positions in determining binding specificity) as these are particularly variable in different ZFPs. Binding to a longer DNA sequence improves the specificity of the zinc finger attaching to its target site and only a DNA sequence having six triplets (or more) is unique in the human genome.
The Web-Server
ZiF-Predict is a web server for predicting recognition helices for C2H2 zinc fingers that bind to specific target DNA sites. This prediction is based on artificial neural network using an exhaustive dataset of zinc finger proteins and their target DNA triplets.
The web-server has a user-friendly interface. Users can select the option for two or three zinc fingers to be predicted in a modular or synergistic fashion for the input DNA sequence. The recognition helices already reported in literature are also made available for the user.
Additionally, it also has an option for the prediction of zinc finger nucleases (ZFNs) that is widely used in targeted genome editing.
The ZiF-Predict network model consists of an input layer followed by two hidden layers and a single output neuron as shown in the figure below.
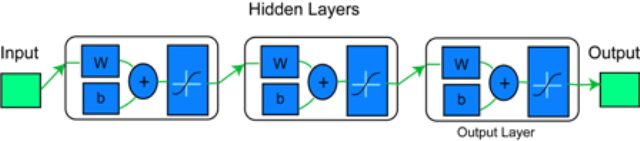
Graphical representation of the ZiF-Predict network model
The Applicability
ZiF-Predict would be valuable for researchers interested in designing specific zinc finger transcription factors and ZFNs for several biological and biomedical applications. This server would be handy for rapid screening of gene-specific ZFP and ZFN target sites in plant or mammalian genomes. This will greatly aid genome researchers to design and/or evolve ZFPs for creating custom zinc finger-chimeric proteins for gene regulation, enzyme engineering, genome editing, gene therapy and several such genomic applications, including human therapeutics.
|
sundar@dbeb.iitd.ac.in |