
|
| |
  |
|
| User Guide |
ZiF-Predict is a web server for predicting recognition helices for C2H2 zinc fingers that bind to specific target DNA sites. This prediction is based on artificial neural network (ANN) using an exhaustive dataset of zinc finger proteins and their target DNA triplets.
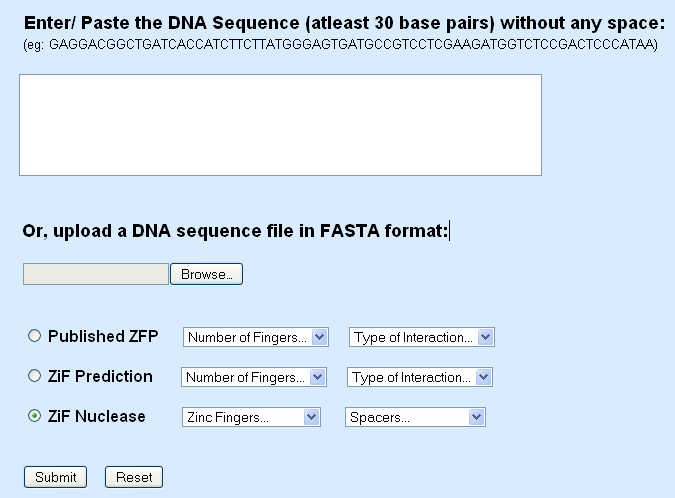
Home Page
Inputting the DNA Sequence
The user can input the DNA sequence, for the prediction of the zinc finger of interest, in two ways –
1. The raw DNA sequence can be pasted in to the space provided (Click in the text box to paste)
2. The DNA sequence can be uploaded as a fasta file which can be uploaded by using the ‘Browse’ button
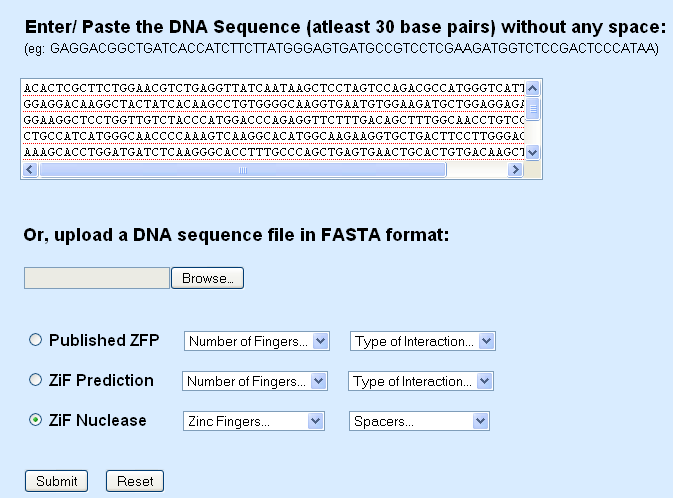
Copied DNA sequence
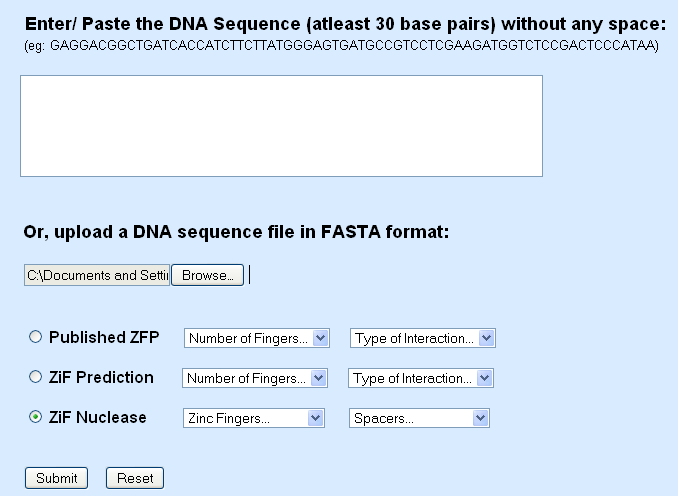
Fasta Format
Options
According to the user's interest, already published data or zinc finger predictions can be pulled out using ZiF-Predict.
I. Published Zinc Finger Proteins
Additionally the user is also empowered with the information whether some zinc finger protein has already been reported in literature by selecting the option of Published ZFPs (i.e. ZFPs that has been experimentally verified in vitro and/or in vivo ) .
II. Prediction
The user is provided different options for predicting ZFP and ZFN using ANN –
1. Prediction of 2 finger ZFPs for the input target DNA considering to be modular entities
2. Prediction of 3 finger ZFPs for the input target DNA considering to be modular entities
3. Prediction of 2 finger ZFPs for the input target DNA keeping in mind the synergistic/dependent effect
4. Prediction of 3 finger ZFPs for the input target DNA keeping in mind the synergistic/dependent effect
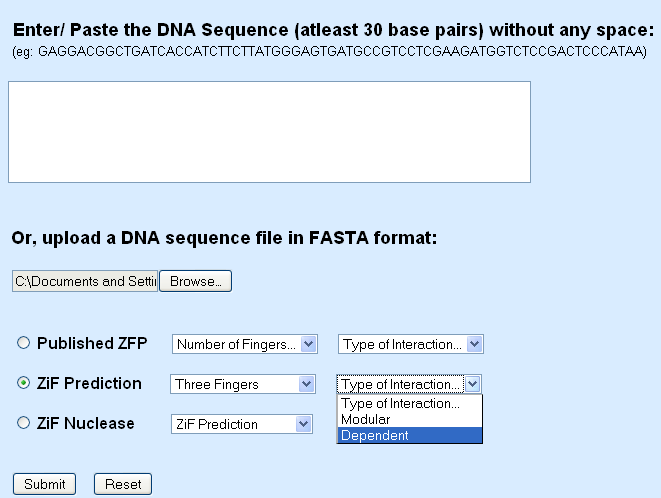
ZF Prediction options
III. Zinc Finger Nucleases
The third choice provided to the user is for Zinc Finger Nucleases (ZFNs). On selecting this option ZiF-Predict identifies a site in the user's input DNA where a nuclease enzyme can possibly bind and also predicts the ZFN recognition helices.
Here again the user is allowed the freedom to chose (by selecting the respective options from the drop-down menu) between
1. Published ZFN sequences
2. Predictions made by ZiF-Predict
A zinc finger nucleases enzyme binds to both the strands, i.e. the forward strand and the reverse strand, of the DNA, mostly separated by a spacer. ZiF-Predict provides the user the liberty to choose amongst spacer length of 4, 5 and 6 (these are tested to be the most favoured spacer lengths for the correct binding and functioning of the ZFN)
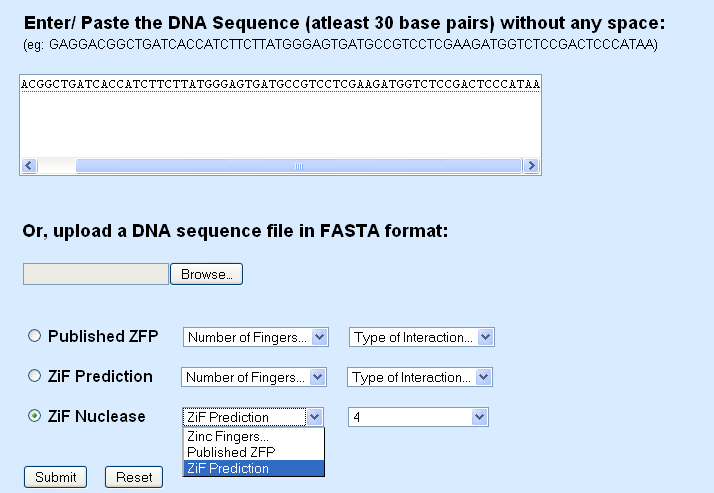
Zinc Finger Nuclease options
Result
After selecting the appropriate options click on the ‘Submit’ button to get the results.
To clear the selcted options and start afresh, click on the ‘Reset’ button. Sample Result pages are shown below :
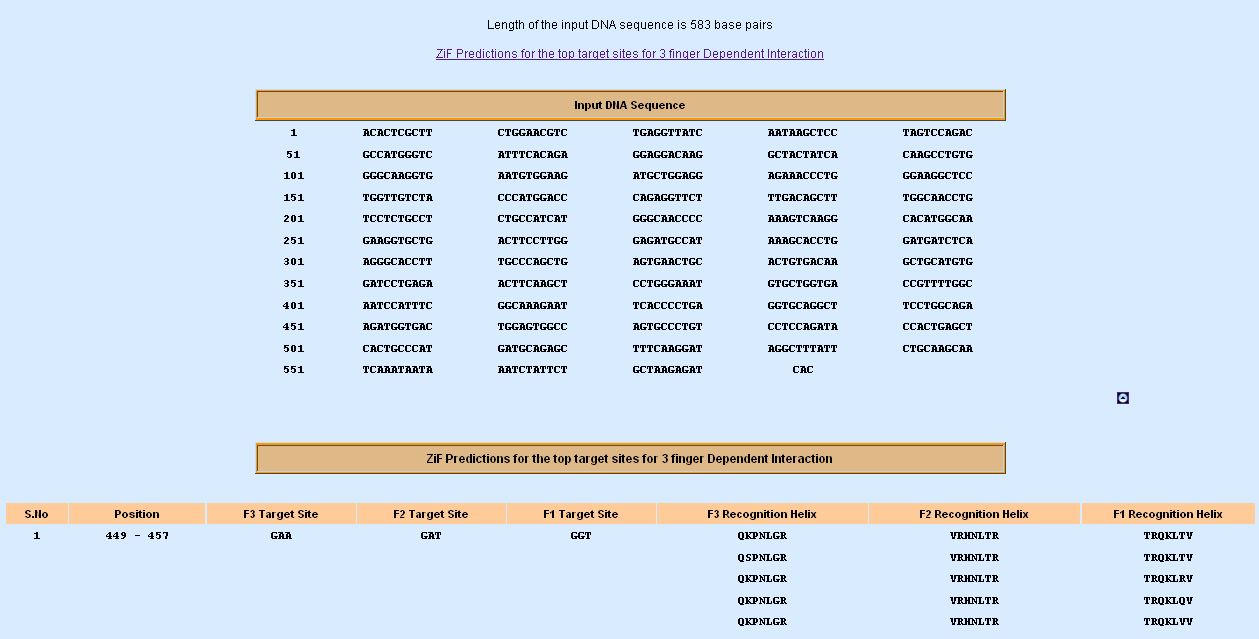
Zinc Finger Predictions - 3 Fingers; dependant
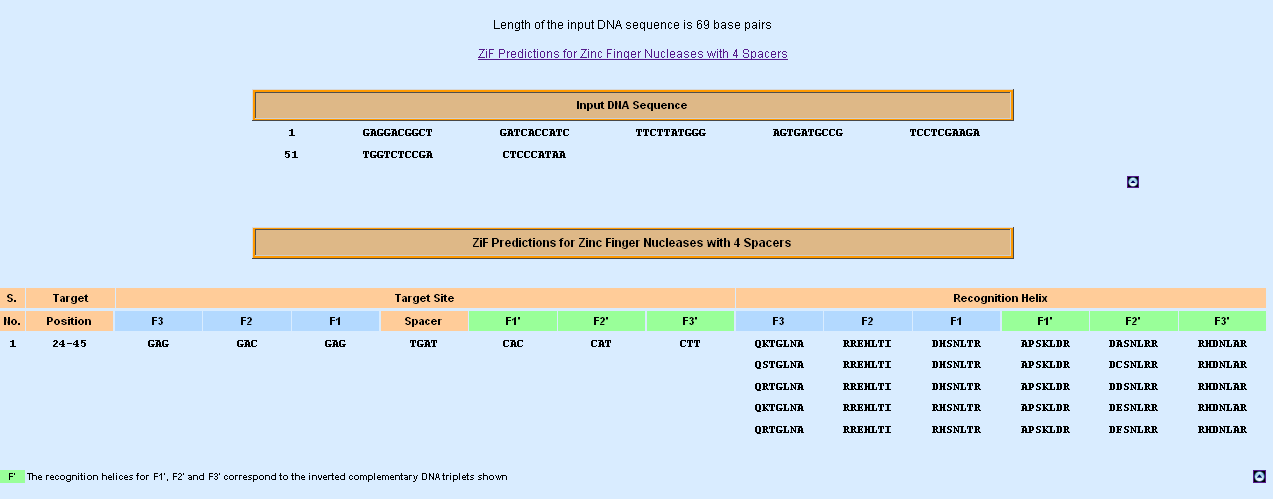
Zinc Finger Nuclease – Zif Prediction; 4 (spacer length)
Queries and suggestions are welcome
 |
|
sundar@dbeb.iitd.ac.in |